Transferring data to and from the cluster
The storage on the cluster isn't generally visible from other machines on site, including your desktop or laptop. If you want to move data on to or off the cluster you will need to do an explicit transfer to some other storage system.
In general we urge people to not duplicate data. The home and group shares on the cluster are secure forms of storage which are automatically backed up so it's perfectly OK to keep your data on there long term. Please also remember that any data you see on /bi/sequencing and /bi/imaging are also backed up, so whilst you can do futher processing of this data in your own areas you don't need to copy data from there.
If you need to move data to/from the cluster then you can do this in one of two ways:
- Using the cluster web interface
- Using an SFTP transfer program
Move data using the cluster web interface
The cluster web interface at https://capstone.babraham.ac.uk/", under the Programs tab, has a FileBrowser program which lets you look at your data, but also upload or download from the cluster.
After starting the file browser in the share you want to work on (home, group or scratch) you can then either select a file or folder in the main view and press the download button in the toolbar to download it.
To upload you can just drag and drop onto the web interface. You can upload either single files, or entire directories this way.
Move data using an SFTP program
Graphical Programs
There are a number of graphical programs which can be used to do data transfer using the SFTP protocol, which is what the cluster uses. Commonly used ones are:
- Cyberduck (Windows or OSX)
- Filezilla (Windows or OSX)
- Bitvise SSH Client (Windows only)
- WinSCP (Windows only)
When setting up a connection in these clients you will either be asked for a connection URL, or for the separate details of the connection.
The connection URL will be:
sftp://[username]@capstone.babraham.ac.uk
For programs which ask for the details separately the information you need will be:
- Hostname:
capstone.babraham.ac.uk - Protocol:
sftp
Username and password are your normal login username and password.
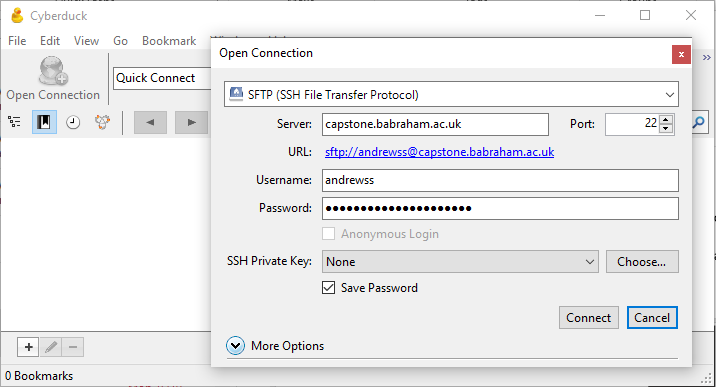
Cyberduck configuration
Select Open Connection from the top left of the program
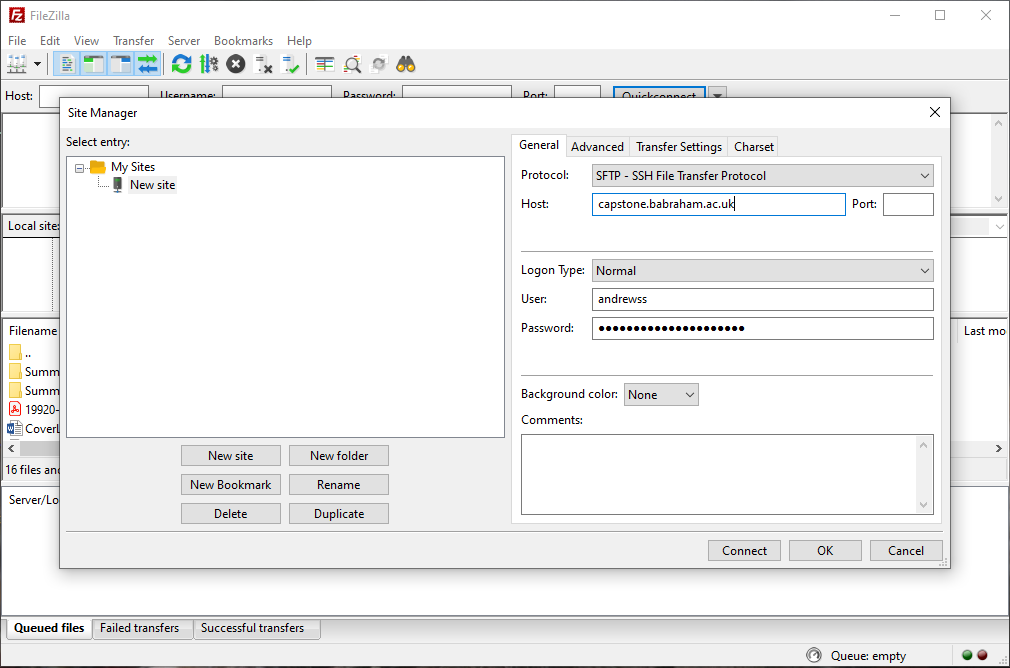
Filezilla configuration
Select File > Site Manager from the main menu
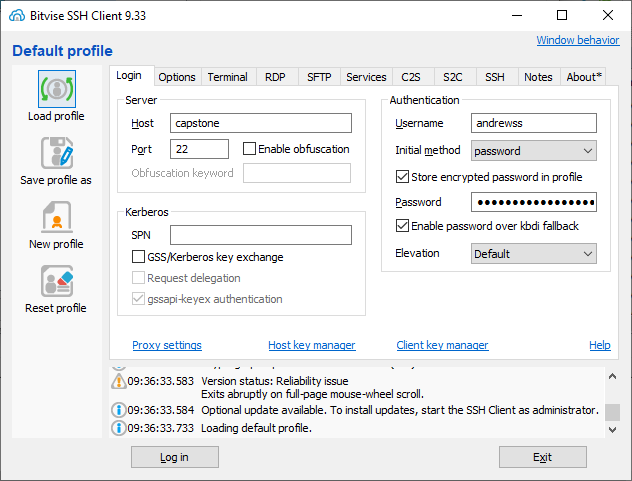
Bitvise SSH configuration
The main connection window should open as soon as the program opens. Once you've made a connection it will stay in your system tray at the bottom right of your screen. You can right click on the bitvise icon in the tray and select New SFTP window and it should open the connection without you having to log in again.
If you get an error saying that the connection failed then there is an additional step you'll need to take

Go into the SSH options
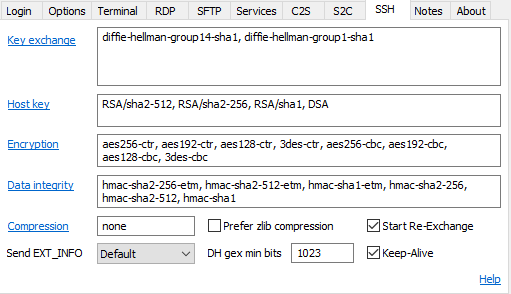
Click on the blue link which says Key exchange and then tick the box which says diffie-hellman-group14-sha256
It should now work. You'll only need to do this once.
WinSCP configuration
Select Tabs > New Tab > Remote Tab
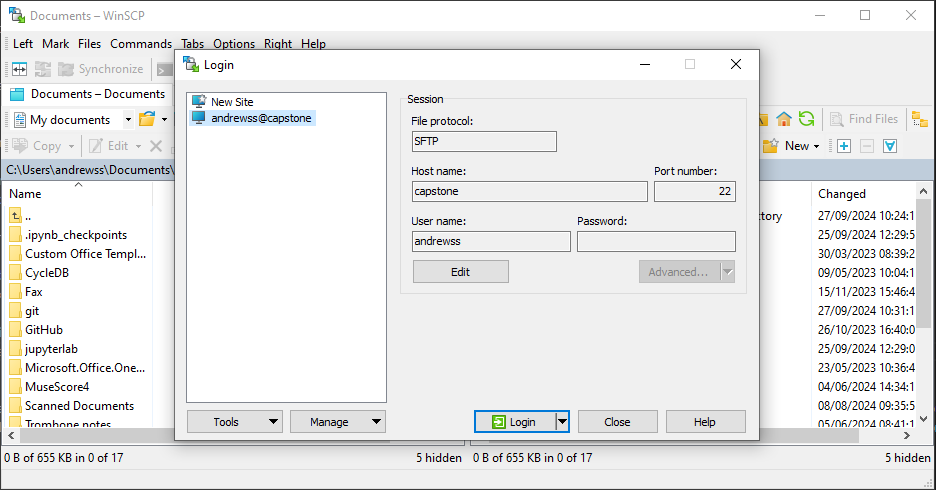
Once you have connected via these programs you will either have a single window from which you can drag and drop data to and from your local machine, or you will have a two panel window where the left side is your local machine and the right is the cluster and you can move data from one side to the other.
Command line transfers
If you want to copy data using a command line interface then you can use a command line sftp program on any platform which supports this.
The command to connect will be:
sftp [username]@capstone.babraham.ac.uk
Once connected you can use commands as shown below:
# List all files in the current directory
sftp> ls
10XCourse
10X_Integration
3_ChIP-Seq
ATAC seq second.smk
Aaron
Angelman
Annotated Probe Report for ID All vs All.txt
...
# Move to a different directory
sftp> cd Temp
# Download a single file
sftp> get fastqc_v0.12.1.zip
Fetching /bi/home/andrewss/Temp/fastqc_v0.12.1.zip to fastqc_v0.12.1.zip
fastqc_v0.12.1.zip
# Download a whole folder
sftp> get -r FastQC/
Fetching /bi/home/andrewss/Temp/FastQC/ to FastQC
Retrieving /bi/home/andrewss/Temp/FastQC
htsjdk.jar 100% 1757KB 3.7MB/s 00:00
README.md 100% 1044 13.9KB/s 00:00
INSTALL.txt 100% 6477 101.2KB/s 00:00
LICENSE 100% 34KB 429.6KB/s 00:00
...
# Download multiple files
sftp> mget *jar
Fetching /bi/home/andrewss/Temp/FastQC/cisd-jhdf5.jar to cisd-jhdf5.jar
cisd-jhdf5.jar 100% 9050KB 5.3MB/s 00:01
Fetching /bi/home/andrewss/Temp/FastQC/htsjdk.jar to htsjdk.jar
htsjdk.jar 100% 1757KB 7.9MB/s 00:00
Fetching /bi/home/andrewss/Temp/FastQC/jbzip2-0.9.jar to jbzip2-0.9.jar
jbzip2-0.9.jar 100% 49KB 872.4KB/s 00:00
# List local files
sftp> lls
cisd-jhdf5.jar connecting.md docs FastQC fastqc_v0.12.1.zip help.md htsjdk.jar introduction.md
# Upload a single file
sftp> put help.md
Uploading help.md to /bi/home/andrewss/Temp/help.md
help.md 100% 0 0.0KB/s 00:00
# Upload multiple files
sftp> mput *jar
Uploading cisd-jhdf5.jar to /bi/home/andrewss/Temp/cisd-jhdf5.jar
cisd-jhdf5.jar 100% 9050KB 2.7MB/s 00:03
Uploading htsjdk.jar to /bi/home/andrewss/Temp/htsjdk.jar
htsjdk.jar 100% 1757KB 2.7MB/s 00:00
Uploading jbzip2-0.9.jar to /bi/home/andrewss/Temp/jbzip2-0.9.jar
jbzip2-0.9.jar
# Upload an entire folder
sftp> put -r FastQC/
Uploading FastQC/ to /bi/home/andrewss/Temp/temp/FastQC
Entering FastQC/
cisd-jhdf5.jar 100% 9050KB 2.8MB/s 00:03
Entering FastQC/Configuration
adapter_list.txt 100% 1355 52.6KB/s 00:00
contaminant_list.txt 100% 13KB 391.9KB/s 00:00
...
# Exit the connection
sftp> bye